
Juexiao Zhou 周觉晓
Tenure-track Assistant Professor @CUHK-Shenzhen
Healthcare Ethical AI Lab, HEAL (意为“治愈”,技术创新助力延长人类寿命)
The Chinese University of Hong Kong, Shenzhen
Email: juexiao.zhou[AT]gmail.com
Office: Zhi Xin Building, 403b, CUHK-Shenzhen
CV (last update: 2026.1) | Github | Linkedln | ORCID | Google Scholar | DBLP
Openings: Remote/Onsite Intern, PhD/MPhil students, Research Assistant
Recent News:
- Jan 2026, one paper is accepted by Nature Methods
- Nov 2025, two innovation awards for Outstanding Scientific Research achievements (第二十七届高交会“优秀科研成果创新奖”) were received at The 27th China Hi-tech fair
- Oct 2025, five GBD 2023 papers are accepted by The Lancet (IF=168.9)
- Oct 2025, one paper is accepted by npj Artificial Intelligence
- Oct 2025, I am happy to serve as the Member of ISCB-China Council
- Oct 2025, Agentic Bioinformatics paper is accepted by Briefings in Bioinformatics (IF=7.7)
- Aug 2025, I have joined CUHK-Shenzhen as an Assistant Professor
- Jun 2025, I am happy to serve as the editor of npj Artificial Intelligence
- May 2025, got my Doctor of Philosophy degree, Ph.D. in Computer Science at King Abdullah University of Science and Technology.
- May 2025, passed my Ph.D. dissertation defense titled "Towards Privacy-preserving Artificial Intelligence (PAI) for Healthcare and Bioinformatics"
- May 2025, I am happy to serve as the co-chair of IS-HIS 2025 Symposium, ICCNS2025, Varna, Bulgaria
- Mar 2025, fair medical LLM paper is accepted by Nature Computational Science (IF=12)
- Feb 2025, I am invited to attend the launching ceremony of The Global AI for Science Developer Community, 2025 Global Developer Conference, Shanghai, China
- Jan 2025, pulmonary artery and vein segmentation paper is accepted by Nature Communications (IF=17)
- Dec 2024, chemical LLM paper is accepted by Communications Chemistry (IF=5.9)
- Nov 2024, I am happy to join the Editorial Board of BMC Bioinformatics
- Nov 2024, We showcased MOSS2.ai at Saudi's first Space Tech Incubation Program
- Sep 2024, GBD 2021 upper respiratory infections is accepted by The Lancet Infectious Diseases (IF=36.4)
- Aug 2024, AutoBA is accepted by Advanced Science (IF=15.1)
- Jul 2024, our recent work SkinGPT-4 is reported by Arab News
- Jun 2024, SkinGPT-4 paper is accepted by Nature Communications (IF=17)
- Jun 2024, a paper is accepted by Bioinformatics (IF=5.8)
- May 2024, GBD 2021 Forecasting Capstone paper is accepted by The Lancet (IF=168.9)
- May 2024, GBD 2021 Risk Factors Capstone paper is accepted by The Lancet (IF=168.9)
- May 2024, a paper is accepted by Computers in Biology and Medicine (IF=7.7)
- Apr 2024, GBD 2021 Global incidence, prevalence, YLDs, DALYs, and HALE paper is accepted by The Lancet (IF=168.9)
- Apr 2024, GBD 2021 Causes of Death paper is accepted by The Lancet (IF=168.9)
- Mar 2024, GBD 2021 Global fertility paper is accepted by The Lancet (IF=168.9)
- Mar 2024, Patient privacy paper is accepted by Trends in Genetics (IF=11.4)
- Mar 2024, GBD 2021 Demographics paper is accepted by The Lancet (IF=168.9)
- Feb 2024, invited to the TV show "Investing Now for Future in Hong Kong and the Middle East:New Cradle of Innovation" by RTHK 香港電台
- Dec 2023, PPML-Omics is accepted by Science Advances (IF=15.4)
- Dec 2023, PPPML-HMI is accepted by Computers in Biology and Medicine (IF=7.7)
- Nov 2023, invited to give a talk to the Saudi Olympiad Elite Camp
- Oct 2023, AFS is accepted by Nature Communications (IF=17)
- Sep 2023, passed my Ph.D. proposal titled "Towards Privacy-preserving Artificial General Intelligence (PAGI) for Healthcare and Bioinformatics"
- Jun 2023, COVID-19 paper is accepted by Nature Communications (IF=17)
- Mar 2023, spatial transcriptomics paper is accepted by Nature Communications (IF=17)
- Feb 2023, invited to give a talk at Rising Stars in AI Symposium 2023 at KAUST
- Oct 2022, DeeReCT-TSS paper is accepted by Genomics, Proteomics & Bioinformatics (IF=10.1)
- May 2022, COVID-19 paper is accepted by Nature Machine Intelligence (IF=27.2)
- Dec 2021, got my master of science degree, M.S. in Computer Science at King Abdullah University of Science and Technology.
Short Bio
周觉晓自2025年8月起担任香港中文大学(深圳)数据科学学院 tenure-track 助理教授。他于2020年获得南方科技大学生物信息学理学学士学位,2025年获得阿卜杜拉国王科技大学计算机科学博士学位。其研究方向聚焦计算机科学与生物医学的交叉领域,主要研究人工智能驱动的智慧医疗、生物信息学,以及临床场景中符合伦理、可信赖的人工智能。技术上关注于研发前沿深度学习模型与大语言模型,用于在各类临床场景中实现疾病检测、预后评估与风险评估。他已在顶级期刊和会议发表40余篇学术论文,包括Nature Methods, Nature Machine Intelligence, Science Advances, Nature Computational Science, Nature Communications, The Lancet, Genome Research, Trends in Genetics, Bioinformatics, IEEE Transactions on Medical Imaging (IEEE TMI), MICCAI等。其中,以第一作者身份发表论文11篇,包括 Nature Methods (2026), Nature Communications (2023, 2023, 2024), Science Advances (2023)等。其研究成果被Arab News, Radio Television Hong Kong (RTHK), Inside Precision Medicine等多家主流媒体报道。他同时是CAAI, APBioNET, GBD的活跃会员。他担任Nature, Nature Methods, Nature Communications, Medical Image Analysis,Genome Biology, Genome Research, NeurIPS, SIGKDD, MICCAI等众多顶级期刊与会议的审稿人并审稿70余次,同时担任BMC Bioinformatics, npj Artificial Intelligence, Biomedical Informatics等期刊的编委。
Juexiao Zhou has been a Tenure-Track Assistant Professor at the School of Data Science, The Chinese University of Hong Kong, Shenzhen, since 2025. He received his Bachelor of Science degree in Bioinformatics from the Southern University of Science and Technology (SUSTech) in 2020 and his Doctor of Philosophy (Ph.D.) degree in Computer Science from King Abdullah University of Science and Technology (KAUST) in 2025. His research spans the intersection of computer science and biomedicine, with a primary focus on AI-driven intelligent healthcare, bioinformatics, and ethical, trustworthy AI in clinical contexts. Juexiao develops cutting-edge deep learning models and large language models (LLMs) to facilitate disease detection, prognosis, and risk assessment across diverse clinical settings. He has authored over 40 publications in top-tier journals and conferences, including Nature Methods, Nature Machine Intelligence, Science Advances, Nature Computational Science, Nature Communications, The Lancet, Genome Research, Trends in Genetics, Bioinformatics, IEEE Transactions on Medical Imaging (IEEE TMI), and MICCAI. Among these, 11 are first-author papers, including contributions to Nature Methods (2026), Nature Communications (2023, 2023, 2024), and Science Advances (2023). His research has been featured by major media outlets such as Arab News, Radio Television Hong Kong (RTHK), and Inside Precision Medicine. He is an active member of the Chinese Association for Artificial Intelligence (CAAI), the Asia Pacific Bioinformatics Network (APBioNET), and GBD. He has reviewed over 70 times for numerous leading journals and conferences, including Nature, Nature Methods, Nature Communications, Medical Image Analysis, Genome Biology, Genome Research, NeurIPS, SIGKDD, and MICCAI. Additionally, he holds editorial positions at BMC Bioinformatics, npj Artificial Intelligence, and Biomedical Informatics.
Education
He got the Ph.D. degree in Computer Science at King Abdullah University of Science and Technology under the supervision of Prof. Xin Gao from Dec 2021 - May 2025.
He got the Master of Science degree, M.S. in Computer Science at King Abdullah University of Science and Technology under the supervision of Prof. Xin Gao in Dec 2021.
He got the Bachelor of Science degree, B.S. (Honored) in Bioinformatics at Southern University of Science and Technology under the supervision of Prof. Wei Chen in Jun 2020.
I said “Hello World” in January 1999 in China.
Research Interests
-
AI-driven Intelligent Healthcare: I develop cutting-edge AI algorithms, particularly DL and LLMs, to enable disease detection, prognosis, and risk assessment across clinical settings.
-
Bioinformatics: I develop intelligent computational frameworks to decode gene regulatory networks, predict protein function and structure, and model complex biological systems. I also develop curiosity-driven AI scientists based on AI agents.
-
Ethical and Trustworthy AI in Healthcare: I investigate the ethical and societal implications of AI in medicine, focusing on pressing challenges such as privacy, security, bias, fairness, toxicity and so on.
Selected Publications
(# equal contribution, * corresponding author) [Full list]
-
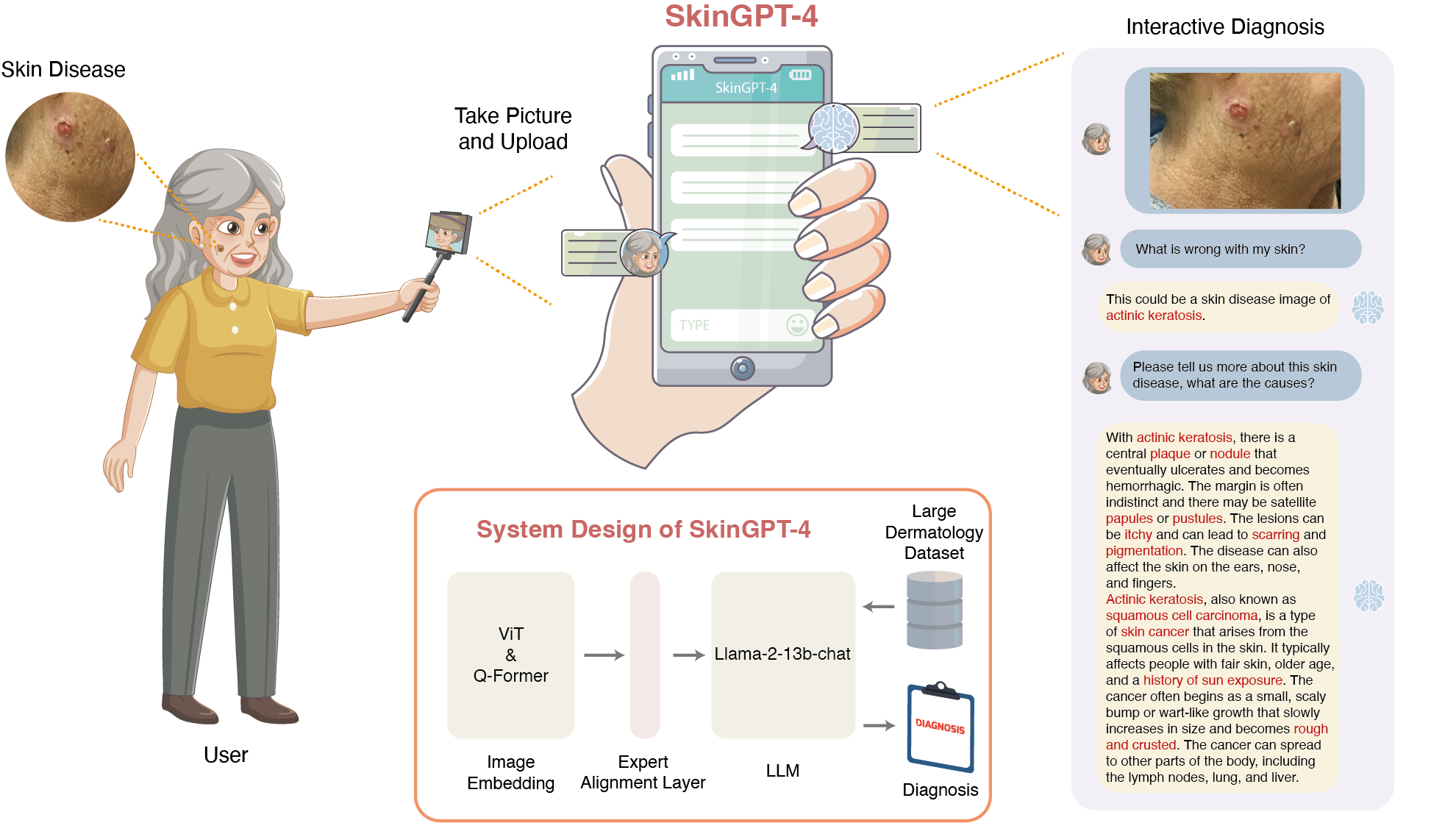
Pre-trained Multimodal Large Language Model Enhances Dermatological Diagnosis using SkinGPT-4.
Juexiao Zhou#, Xiaonan He#, Liyuan Sun#, Jiannan Xu, Xiuying Chen, Yuetan Chu, Longxi Zhou, Xingyu Liao, Bin Zhang, Shawn Afvari, Xin Gao*
Nature Communications
DOI: https://doi.org/10.1038/s41467-024-50043-3
Access: online, pdf
Press: Arab News, ChicHue, KAUST Discovery, 转化医学网 -
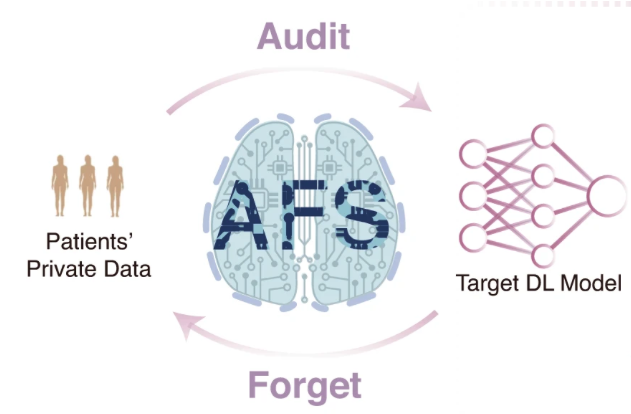
A unified method to revoke the private data of patients in intelligent healthcare with audit to forget.
Juexiao Zhou#, Haoyang Li#, Xingyu Liao, Bin Zhang, Wenjia He, Zhongxiao Li, Longxi Zhou, Xin Gao*
Nature Communications
DOI: 10.1038/s41467-023-41703-x
Access: online, pdf
Press: KAUST Discovery, Enerzine -
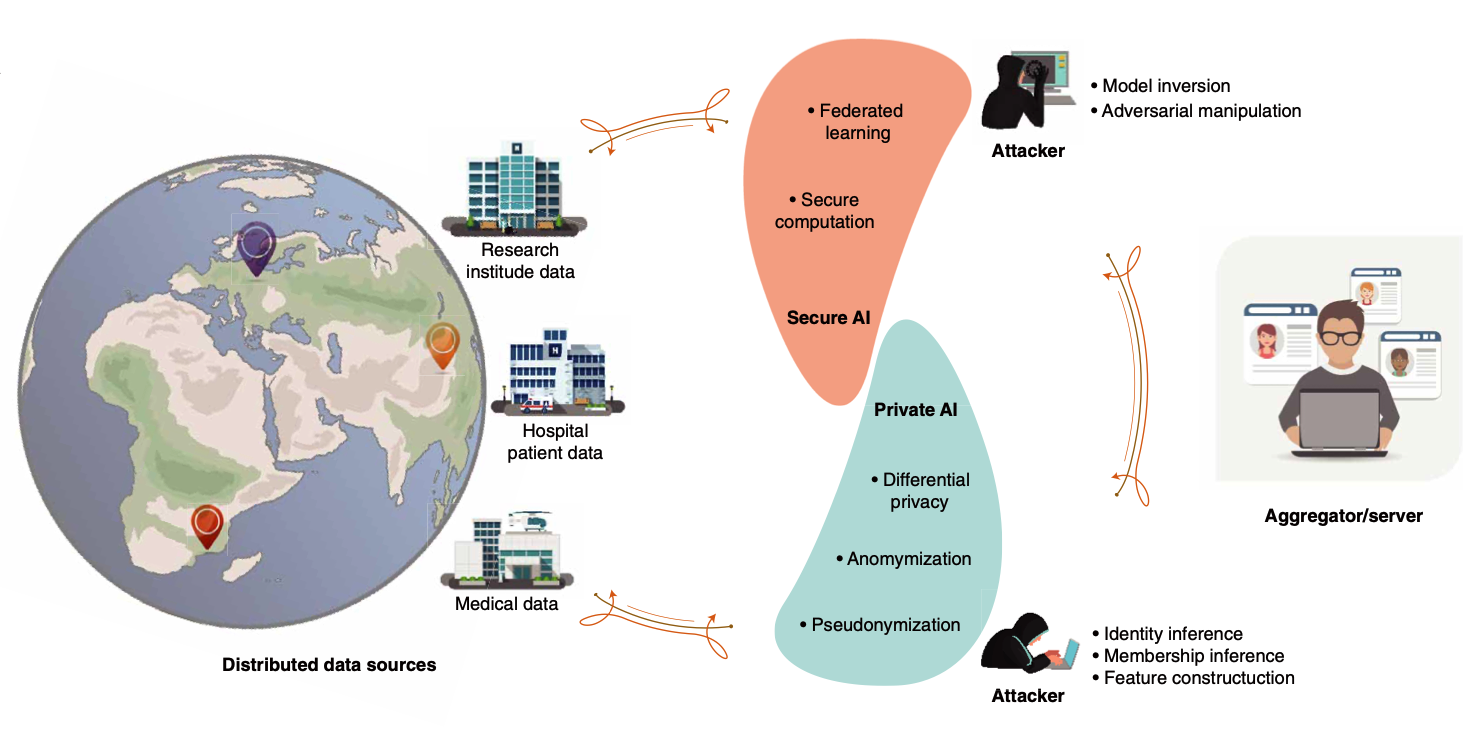
PPML-Omics: A privacy-preserving federated machine learning method protects patients’ privacy in omic data.
Juexiao Zhou#, Siyuan Chen#, Yulian Wu#, Haoyang Li, Bin Zhang, Longxi Zhou, Yan Hu, Zihang Xiang, Zhongxiao Li, Ningning Chen, Wenkai Han, Di Wang and Xin Gao*.
Science Advances
DOI: 10.1126/sciadv.adh8601
Access: online, pdf
Press: Inside Precision Medicine, Today Headline, Tech Xplore, ScienMag, Bioengineer.org, Newswise, EurekAlert, Biocompare, nabd.ws -
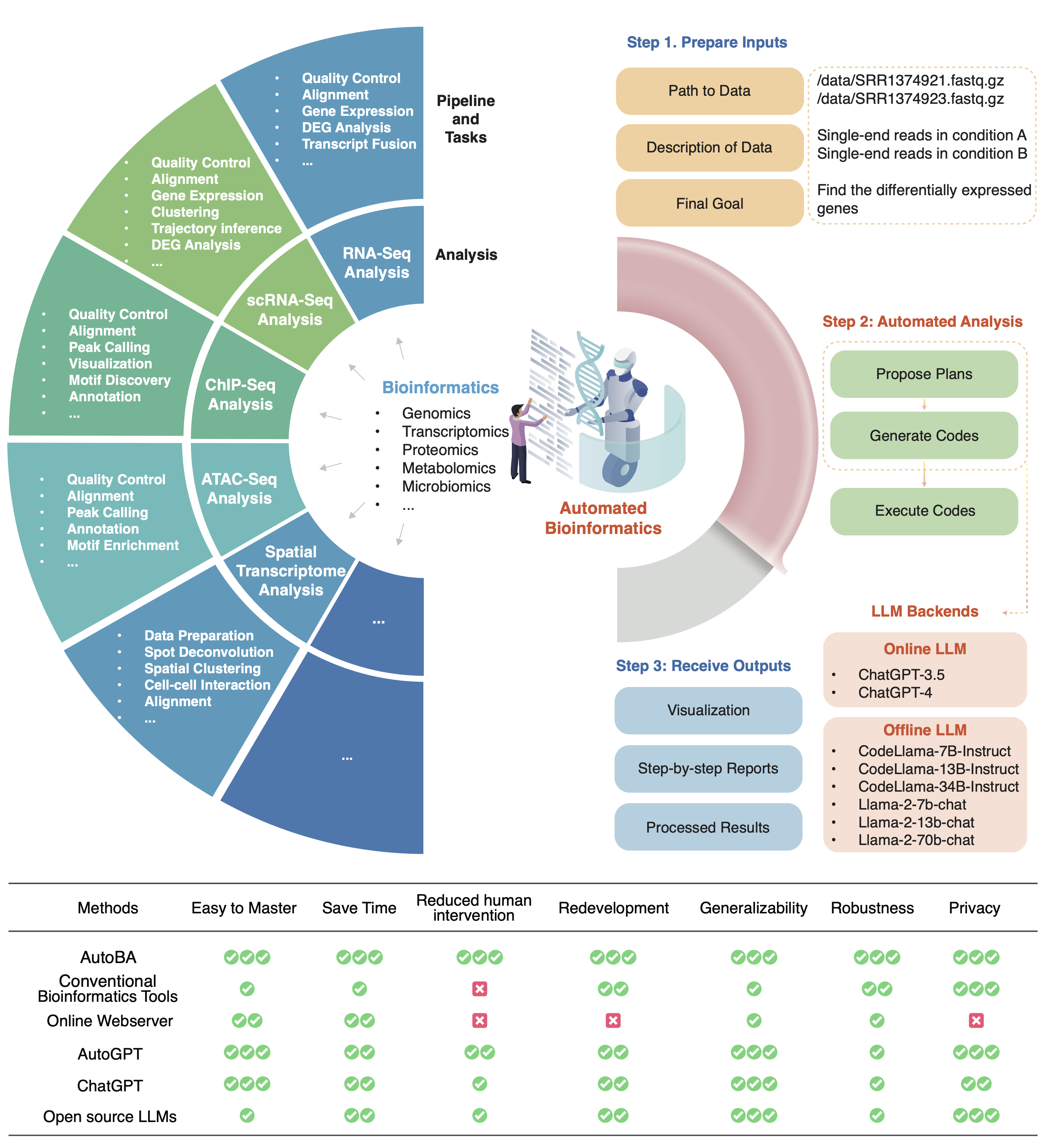
An AI Agent for Fully Automated Multi-omic Analyses.
Juexiao Zhou#, Bin Zhang#, Guowei Li, Xiuying Chen, Haoyang Li, Xiaopeng Xu, Siyuan Chen, Liwei Liu, Xin Gao*
Advanced Science
DOI: https://doi.org/10.1002/advs.202407094
Access: online, pdf
Press: CBIRT news, Medium -
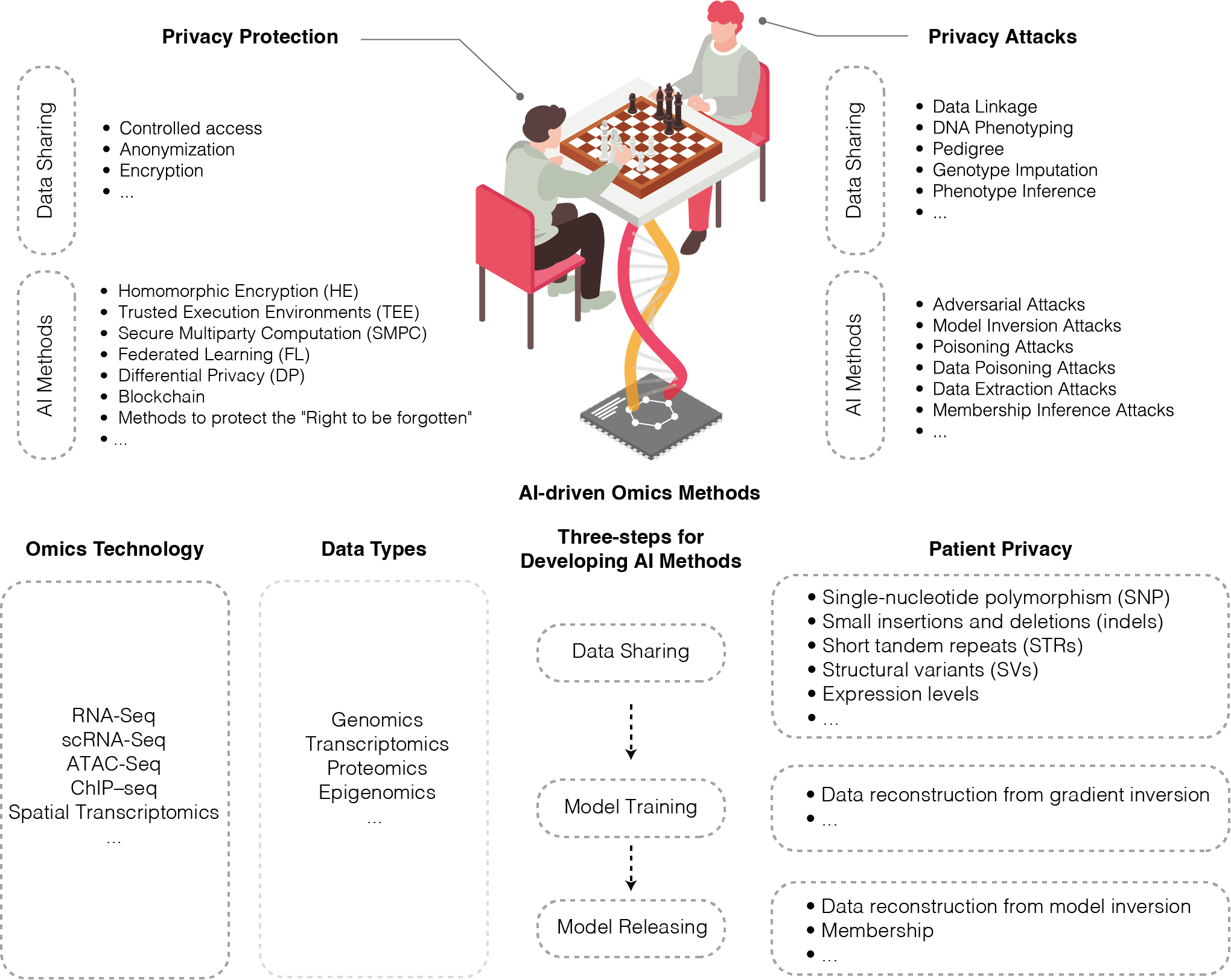
Patient privacy in AI-driven omics methods.
Juexiao Zhou, Chao Huang, Xin Gao
Trends in Genetics
DOI: https://doi.org/10.1016/j.tig.2024.03.004
Access: online, pdf -
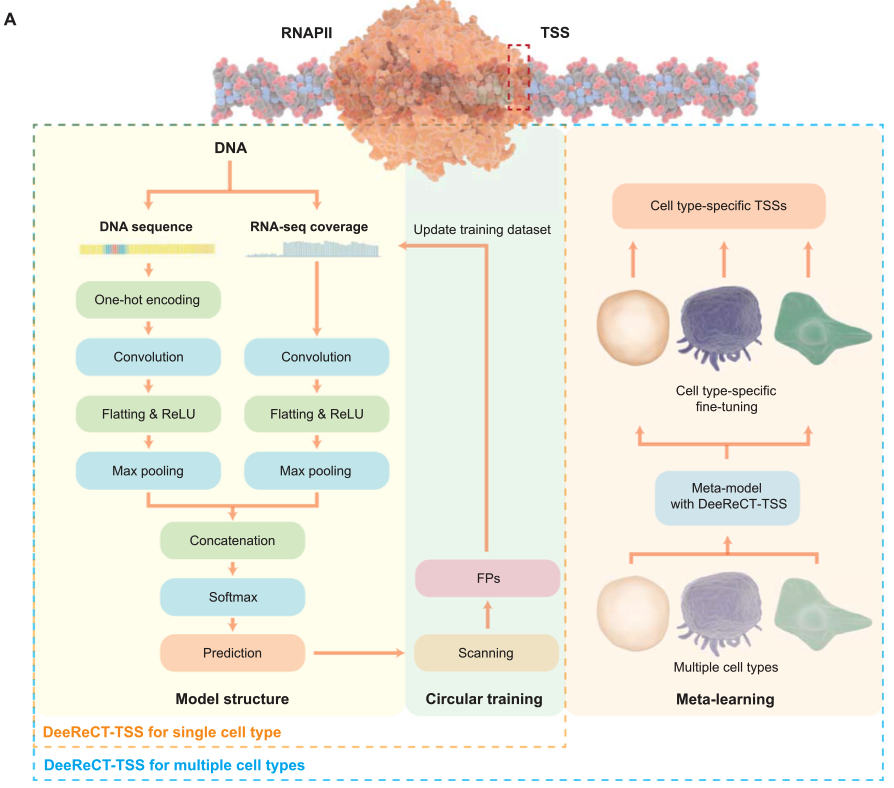
Annotating TSSs in Multiple Cell Types Based on DNA Sequences and RNA-seq Data via DeeReCT-TSS.
Juexiao Zhou#, Bin Zhang#, Haoyang Li, Longxi Zhou, Zhongxiao Li, Yongkang Long, Wenkai Han, Mengran Wang, Huanhuan Cui, Wei Chen, Xin Gao*.
Genomics, Proteomics & Bioinformatics
DOI: 10.1016/j.gpb.2022.11.010
Access: online, pdf -
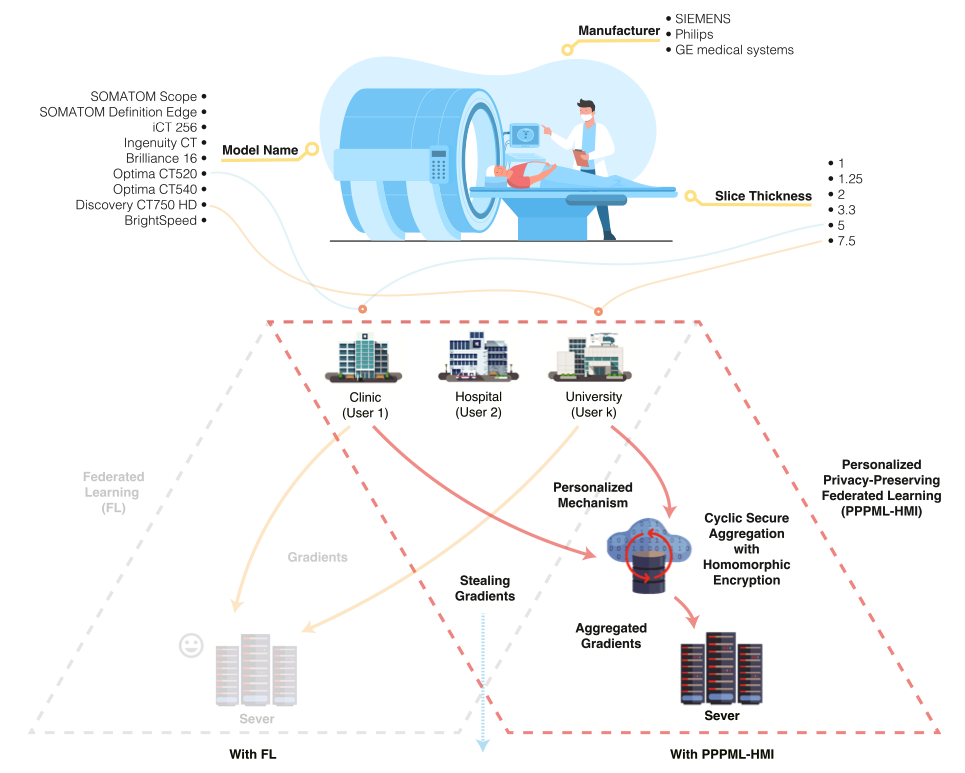
Personalized and privacy-preserving federated heterogeneous medical image analysis with PPPML-HMI.
Juexiao Zhou#, Longxi Zhou#, Di Wang, Xiaopeng Xu, Haoyang Li, Yuetan Chu, Wenkai Han, Xin Gao*
Computers in Biology and Medicine
DOI: 10.1016/j.compbiomed.2023.107861
Access: online, pdf -
A comprehensive benchmarking with practical guidelines for cellular deconvolution of spatial transcriptomics.
Haoyang Li#, Juexiao Zhou#, Zhongxiao Li, Siyuan Chen, Xingyu Liao, Bin Zhang, Ruochi Zhang, Yu Wang, Shiwei Sun, Xin Gao*
Nature Communications
DOI: 10.1038/s41467-023-37168-7
Access: online, pdf
Press: BioMed, CBIRT news, KAUST news
Chosen as Editors’ Highlights in Nature Communications -
An Interpretable Deep Learning Workflow for Discovering Sub-Visual Abnormalities in CT Scans of COVID-19 Inpatients and Survivors.
Longxi Zhou#, Xianglin Meng#, Yuxin Huang#, Kai Kang#, Juexiao Zhou, Yuetan Chu, Haoyang Li, Dexuan Xie, Jiannan Zhang, Weizhen Yang, Na Bai, Yi Zhao, Mingyan Zhao, Guohua Wang, Lawrence Carin, Xigang Xiao, Kaijiang Yu, Zhaowen Qiu, Xin Gao*.
Nature Machine Intelligence
DOI: 10.1038/s42256-022-00483-7
Access: online, pdf -
Evaluating and mitigating bias in AI-based medical text generation.
Xiuying Chen, Tairan Wang, Juexiao Zhou, Zirui Song, Xin Gao, Xiangliang Zhang
Nature Computational Science
DOI: 10.1038/s43588-025-00789-7
Access: online, pdf
Academia Experience
- Tenure-track Assistant Professor in Computer Science, The Chinese University of Hong Kong, Shenzhen, Aug 2025 - Present
- Student Ambassador, CEMSE, King Abdullah University of Science and Technology, Dec 2021 - Apr 2025
Industry Experience
-
Co-founder & CEO, MOSS.ai, Sep 2024 - Dec 2025
-
Co-founder & Chief Scientist, BeautyX.ai, Sep 2024 -
-
Co-founder & Chief AI Scientist, DermAssure.ai, April 2024 -
Teaching Experience
-
CSC3100 Data Structures, Fall 2025, CUHK-Shenzhen, China
- Position: Instructor
-
DDA3020 Machine Learning, Fall 2025, CUHK-Shenzhen, China
- Position: Instructor
-
CS220, Data Analytics - AI Camp for Ministry of Interior, Fall 2024/2025, Saudi Arabia
- Cooperated with Prof. Xin Gao
- Position: Teaching Assistant
-
CS220, Data Analytics, Fall 2024/2025, KAUST
- Cooperated with Prof. Xin Gao
- Position: Teaching Assistant
-
CS220, Data Analytics - AI Camp for Ministry of Interior, Fall 2023/2024, Saudi Arabia
-
Cooperated with Prof. Xin Gao
-
Position: Senior Teaching Assistant
-
-
CS220, Data Analytics, Fall 2022/2023, KAUST
- Cooperated with Prof. Xin Gao
- Position: Teaching Assistant
-
BioE 201/230 Foundations of Bioengineering, Fall 2022/2023, KAUST
- Cooperated with Prof. Xin Gao
- Position: Guest Lecturer, Building 9 Room 4225
- Contents:
- Lab 1. Genome data analysis, 2022.08.30, colab
- Python basics
- D/RNA sequence analysis
- Biopython
- BLAST
- Reference genome
- Lab 2. Protein sequence analysis, 2022.09.06, colab
- Protein sequence analysis
- MSA
- PDB
- PyMOL
- Secondary structure prediction
- Lab 3. Protein structure and function, 2022.09.13, colab
- Ab initio with PyRosetta
- AlphaFold2
- NucleicNet
- Docking with Smina
- Pfam annotation with deep learning
- Lab 1. Genome data analysis, 2022.08.30, colab
-
CS398 Graduate Seminar, Spring 2022, KAUST
- Cooperated with Prof. Dominik Michels
- Position: Teaching Assistant
Mentoring
I’m very glad to work and grow together with these excellent students.
Honors & Awards
The Chinese University of Hong Kong, Shenzhen
CUHK-Shenzhen, 2025 - Present
Tenure-track Assistant Professor
- Innovation awards for Outstanding Scientific Research Achievements (第二十七届高交会“优秀科研成果创新奖”), The 27th China Hi-tech fair (two awards), 2025
- The first batch of Members of ISCB-China Council, 2025
King Abdullah University of Science and Technology
KAUST, 2021-2025
PhD, Computer Science
GPA: 4.00 / 4.00
- CEMSE Dean’s List Award, 2025
- CEMSE Dean’s List Award, 2024
- CEMSE Dean’s List Award, 2023
- KAUST AII’s NeurIPS travel grant, 2022
- CEMSE Dean’s List Award, 2022
- Excellent Research Award, CEMSE, 2021
King Abdullah University of Science and Technology
KAUST, 2020-2021
Master of Science, Computer Science
GPA: 3.95 / 4.00
- Student ambassador, CEMSE, 2021
- Yearly best student award, CEMSE, 2021
- Full scholarship for MS/PhD study, 2020
Southern University of Science and Technology
SUSTech, 2016-2020
Bachelor of Science, Bioinformatics, Biology
GPA: 3.92 / 4.00 (Top 0.1%), Core GPA: 3.94/4.00
- Outstanding graduate of SUSTech, 2020.
- Cum Laude Graduate of the Department of Biology (Top 1/10), 2020.
- The Guinness world record for “the most vows received by a single civilized act activity”, 2019
- Candidate for 2019 National Scholarship, 2019
- Summer social practice excellent experience Award, 2018
- Excellent Student, The First Prize Scholarship, 2018
- Outstanding volunteer for the 12th CBIS Biennial Meeting, Shenzhen, China, 2018
- Candidate for 2018 National Scholarship, 2018
- Outstanding Volunteer of the 3rd Shenzhen International Life Science & Health Industry Summit (2016)
- Excellent Student, The First Prize Scholarship, 2017
- Dean scholarship, The First Prize Scholarship, 2017
- Excellent Student Cadre, 2017
- Outstanding Volunteer of 2017 Shenzhen International Precision Medicine Summit, 2017
- Alma mater practice excellent team, 2017
- National Literary Creation Award, 2016
- Excellent Student, The Second Prize Scholarship, 2016
Memberships
- Chinese Association for Artificial Intelligence (CAAI) Member, 2018-Present
- The international Asia-Pacific Bioinformatics Network (APBioNET) Member, 2022-Present
- Global Burden of Disease (GBD) Collaborator Network Member, 2023-Present
- CCF生物信息专委会通讯委员, 2025-Present
- Member of ISCB-China Council, 2025-Present
- 广东省医学会肿瘤影像与大数据分会委员, 2025-Present
Press Coverage
- SkinGPT-4 was covered by:
- How a Saudi university is using AI to transform the diagnosis and treatment of skin diseases, Arab News
- How Saudi Arabia’s SkinGPT-4 is Revolutionizing Skin Care?, ChicHue
- 皮肤诊断“智能”升级:阿卜杜拉国王科技大学团队引入多模态大语言模型, 澎湃新闻
- AI tool for diagnosing skin disorders, KAUST Discovery
- PPML-Omics protects patients’ privacy in omic data was covered by:
- Algorithm Proposed to Protect Patient Privacy, Inside Precision Medicine
- An integrated shuffler optimizes the privacy of personal genomic data used for machine learning, Today Headline
- An integrated shuffler optimizes the privacy of personal genomic data used for machine learning, Tech Xplore
- Shuffling the deck for privacy, ScienMag
- Shuffling the deck for privacy, Bioengineer.org
- Shuffling the deck for privacy, Newswise
- Shuffling the deck for privacy, EurekAlert
- Novel Privacy-Preserving Machine-Learning Method Developed for Genomics Data, Biocompare
- نهج ثوري يحافظ على خصوصية البيانات الطبية, nabd.ws
- Revolutionizing Healthcare: Secure Multi-Party Computation and Privacy-Preserving Machine Learning, BNN Breaking
- An integrated shuffler optimizes the privacy of personal genomic data used for machine learning, Microsoft Start
- Shuffling The Deck For Privacy, Eurasia Review
- Audit to forget was covered by:
- Un logiciel pour garantir le droit à l’oubli des patients dans l’IA en milieu de santé, Enerzine
- Safeguarding the right to be forgotten, KAUST Discovery
Invited Talks
- [2025/11/28] 迈向可信赖、强解释性与道德的智慧医疗人工智能(Towards Trustworthy, Explainable and Ethical Intelligent Healthcare)
- Oral. “智汇湾区,数创未来”–2025智慧医疗与数字健康创新融合发展大会 (“Intelligence Empower Digital Future” - 2025 Smart Healthcare and Digital Health Innovation Development Conference), Shenzhen, China.
- [2025/08/10] 迈向智慧医疗的伦理人工智能(Towards Healthcare Ethical AI)
- Oral. 第十届中国计算机学会生物信息学会议, 海南海口, China.
- [2025/07/14] 迈向智慧医疗的伦理人工智能(Towards Healthcare Ethical AI)
- Oral. “新医科的创新实践:人工智能赋能科学研究”, 汕头大学医学院, China.
- [2025/06/06] 迈向智慧医疗的伦理人工智能(Towards Healthcare Ethical AI)
- Oral. BAAI Conference, 2025 (北京智源大会), Beijing, China.
- [2025/05/15] Pre-trained Multimodal Large Language Model Enhances Dermatological Diagnosis using SkinGPT-4.
- Poster. Inaugural KCSH Conference: Shaping the Future of Smart Health, Jeddah, Saudi Arabia.
- [2025/02/24] Private Artificial Intelligence (PAI) for Healthcare.
- Oral. Shanghai Innovation Institute, China.
- [2024/11/27] Pre-trained Multimodal Large Language Model Enhances Dermatological Diagnosis using SkinGPT-4.
- Poster. 1st International Health Conference on Quality and Organization Excellence, Jeddah, Saudi Arabia. certificate
- [2024/11/04] Private Artificial Intelligence (PAI) for Healthcare.
- Oral. Westlake University, China.
- [2024/10/28] Private Artificial Intelligence (PAI) for Healthcare.
- Oral. CUHK-Shenzhen, China.
- [2024/10/22] Pre-trained Multimodal Large Language Model Enhances Dermatological Diagnosis using SkinGPT-4.
- Poster. Health Innovation Week 2024, Global Health Exhibition, Riyadh, Saudi Arabia. certificate
- [2024/08/15] AI for Healthcare: Pre-trained Multimodal Large Language Model Enhances Dermatological Diagnosis using SkinGPT-4.
- Oral. 汕头大学医学院“智能计算赋能医学”, 中国
- [2023/11/22] Introduction to ChatGPT and Application of GPT in Healthcare.
- Oral. Saudi Olympiad Elite Camp, King Abdullah University of Science and Technology, Saudi Arabia
- [2023/09/20] A Unified Method to Revoke the Private Data of Patients in Intelligent Healthcare with Audit to Forget.
- Oral. CBRC Seminar, King Abdullah University of Science and Technology, Saudi Arabia
- [2023/06/12] SkinGPT-4: An Interactive Dermatology Diagnostic System with Visual Large Language Model.
- Oral. Unlocking the Power of “ChatGPT”, King Abdullah University of Science and Technology, Saudi Arabia
- [2023/03/02] Audit to Forget: A Unified Method to Revoke Patients’ Private Data in Intelligent Healthcare.
- Poster. CBRC session, KAUST Research Open Week, Saudi Arabia
- [2023/02/02] Audit to Forget: A Unified Method to Revoke Patients’ Private Data in Intelligent Healthcare.
- Oral & Poster. Spotlight, Rising Stars in AI Symposium 2023 at KAUST, Saudi Arabia
- [2022/11/10] Privacy in Bioinformatics and Intelligent Healthcare.
- Poster. Smart-Health Student Research Symposium, KAUST, Saudi Arabia
- [2022/03/29] PPML-Omics: a Privacy-Preserving federated Machine Learning system protects patients’ privacy from omic data
- Oral. CBRC Dual Seminar, King Abdullah University of Science and Technology, Saudi Arabia
- [2022/03/09] PPML-Omics: a Privacy-Preserving federated Machine Learning system protects patients’ privacy from omic data
- Oral. BDAI重点实验室研究生沙龙第19期, 中国人民大学高瓴人工智能学院,中国北京
Academic Services
Reviewer
| Journal/Conference | # Paper Reviewed |
|---|---|
| AI4D3 @ NeurIPS 2023 | 1 |
| BMC Artificial Intelligence | 1 |
| BMC Bioinformatics | 2 |
| BMC Medical Informatics and Decision Making | 1 |
| Computers in Biology and Medicine | 21 |
| Computational Biology and Chemistry | 3 |
| Computerized Medical Imaging and Graphics | 2 |
| Computational and Structural Biotechnology Journal | 1 |
| Genome Biology | 1 |
| Genome Research | 2 |
| Heliyon | 1 |
| IJCAI-ECAI 2022 | 1 |
| ICONIP 2020 | 1 |
| ICMLA 2021 | 1 |
| ICLR 2026 CAO Workshop | 2 |
| ISMB 2026 | 5 |
| International Journal of Intelligent Systems | 1 |
| IEEE Transactions on Knowledge and Data Engineering | 1 |
| International Journal of Machine Learning and Cybernetics | 1 |
| Journal of Bioinformatics and Computational Biology | 1 |
| Knowledge and Information Systems | 1 |
| Medical Image Analysis | 2 |
| MICCAI 2020 | 1 |
| MICCAI 2024 | 5 |
| Multimedia Systems | 1 |
| Nature | 1 |
| Nature Methods | 3 |
| Nature Communications | 3 |
| npj Precision Oncology | 1 |
| npj Digital Medicine | 2 |
| npj Artificial Intelligence | 1 |
| Quantitative Biology | 1 |
| SIGKDD 2022 | 2 |
| Scientific Reports | 4 |
Editor
- Editorial Board Member of npj Artificial Intelligence
- Editorial Board Member of BMC Bioinformatics
- Guest Editor Assistant in Biomedical Informatics (Special issue: spatial transcriptomics)
- PC member of ISMB 2026
Conference Administrator
- Co-Chair, IS-HIS 2025 Symposium, ICCNS2025, Varna, Bulgaria
- Administrator, The 21st International Conference on Bioinformatics (InCoB2022), certificate
