[NMI] A geometric deep learning approach to predict binding conformations of bioactive molecules 解读
Title: A geometric deep learning approach to predict binding conformations of bioactive molecules
DOI: https://doi.org/10.1038/s42256-021-00409-9
INFO: NATURE MACHINE INTELLIGENCE | VOL 3 | DECEMBER 2021
发表周期: Received: 18 May 2021; Accepted: 28 September 2021; Published online: 2 December 2021
药物设计是近年来AI应用的极火领域之一,其中一个最有挑战的问题就是空间结构设计。
One of the difficulties arises from the fact that only a small portion of the large chemical space will bind to a specific biological target and result in a therapeutic effect.
这篇文章中作者应用深度学习模型DeepDock捕获protein-ligand complex中的原子级微环境特征学习其中的结合构象。
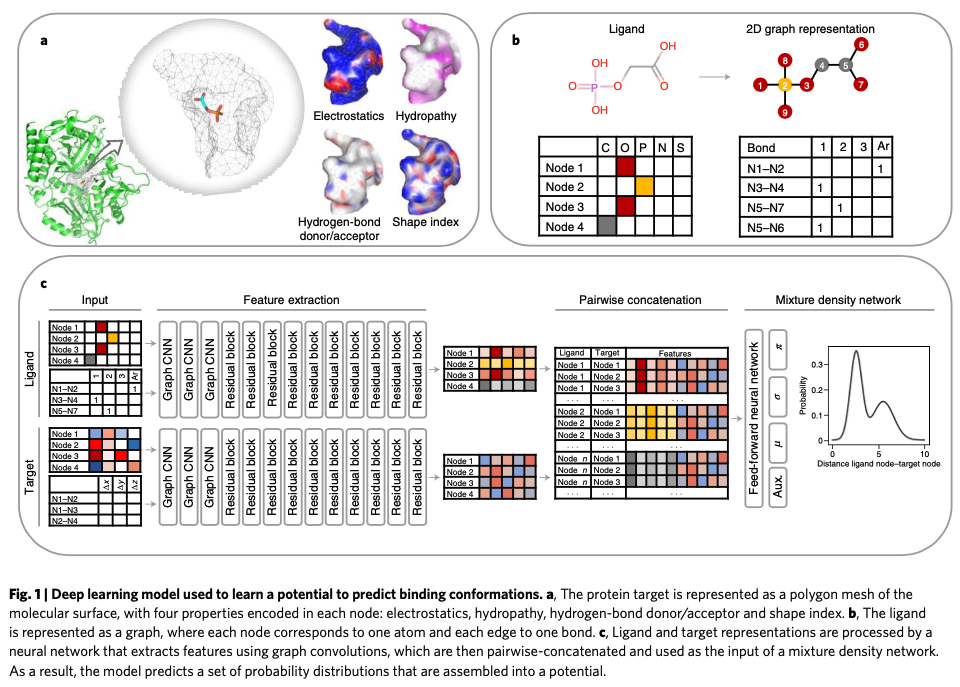
数据集制备
The model reported in this study was trained using the general set of the PDBbind database (v.2019)30, which contains a collection of 17,679 protein–ligand structures with their respective potency (half-maximum inhibitory concentration, dissociation constant and so on). From these, we removed those complexes that are included in the CASF-2016 benchmark and those that failed during the preprocessing step, leaving a total of 16,367 protein–ligand complexes, which were randomly divided in a training set containing 15,000 complexes and a test set with 1,367.
PDBbind数据库(http://www.pdbbind.org.cn)是基于PDB数据库,整合了大量实验统计所得的蛋白亲和力数据。
The PDBbind database is a comprehensive collection of experimentally measured binding affinity data (Kd, Ki, and IC50) for the protein-ligand complexes deposited in the Protein Data Bank (PDB).[1][2] It thus provides a link between energetic and structural information of protein-ligand complexes, which is of great value to various studies on molecular recognition occurred in biological systems.
作者使用图结构表示蛋白空间结构,node表示原子,edge表示bond。
Every node is represented by a one-hot vector that indicates the atom type among 28 possibilities (Be, B, C, N, O, F, Mg, Si, P, S, Cl, V, Fe, Co, Cu, Zn, As, Se, Br, Ru, Rh, Sb, I, Re, Os, Ir, Pt and Hg).
目标蛋白表面使用MSMS建模。
As in MaSIF, protein surfaces were triangulated using MSMS37 with a density of 3.0 nodes per Å^2 and a probe radius of 1.5 Å. The resulting meshes were downsampled to a resolution of 1 Å and processed using PyMesh.
只有具备结合位点性质的node才用于模型训练。
网络训练
如Fig1c,平行的图卷积网络被用来分别提取ligand和target结构中的表征以及特征编码。
Our approach directly uses the molecular surface of the binding site in the form of a polygon mesh.
Both the target mesh and the ligand graph are processed by independent residual GNNs. Through this procedure, the processed node fea- tures not only contain information of an individual atom or point in the molecular surface, but also have information about the other nodes around them. In other words, the processed atom features encode all the atomic environment around a specific atom, whereas the target features encode a patch of the molecular surface around a specific point.
GNN从ligand和target中分别提取出来的特征在特征向量上简单拼接后被送入mixture density network (MDN)。
by using this probability density function we can estimate the likelihood of finding ligand node i separated from a target node j by any distance $d_{ij}$.
模型评估
作者通过在CASF-2016 benchmark预测
- Docking power
- Forward screening power
- Reverse screening power
来评估模型性能。
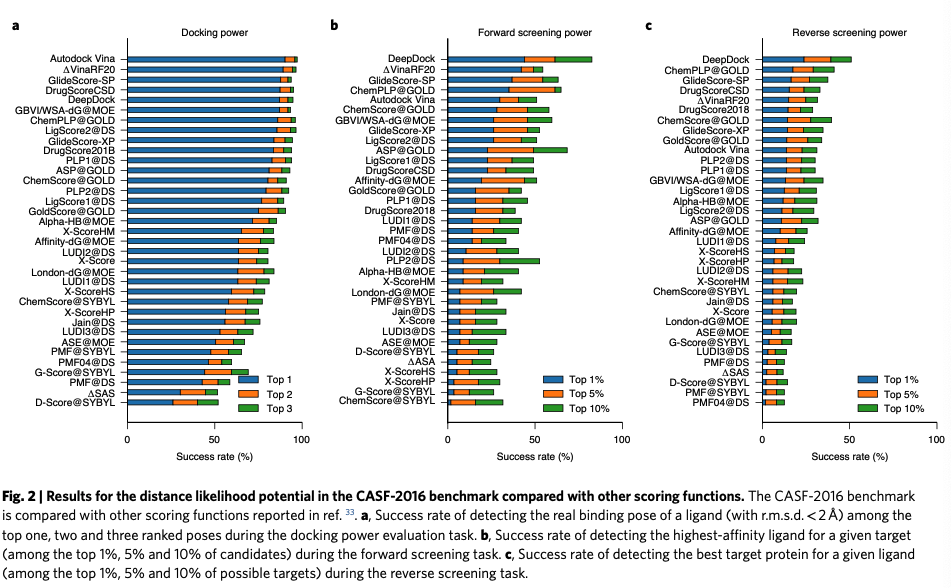
Docking power
The evaluation of docking power measures the ability of a scor- ing function to identify native ligand binding poses among a set of decoys.
Screening power
The evaluation of screening power in CASF-2016 is designed to measure the ability of a scoring function to identify true binders of a specific target from a pool of random compounds.
模型细节
模型可分为3部分:
- 特征提取
- 特征融合
- MDN
First, the node and edge features are projected to a 128D embedding using a linear layer.
We then use a sequence of three GNNs to update each node and edge based on their neighbouring nodes and the type of edges connecting them.
After the initial processing by the GNNs, the node and edge features were processed by 10 residual GNN blocks.
The MDN uses an MLP to create a hidden representation $h_{r,s}$ that combines the concatenated target and ligand node information.
All MLPs used are composed of a linear layer followed by batch normalization and an exponential linear unit (ELU) as activation function.